|
Review Article
New Delhi Metallo-β-lactamase among clinical isolates in Nepal
1 Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu, Nepal
2 Saint Xavier’s College, Maitighar, Kathmandu, Nepal
Address correspondence to:
Surya Prasad Devkota
Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu,
Nepal
Message to Corresponding Author
Article ID: 100010M08SD2018
Access full text article on other devices

Access PDF of article on other devices

How to cite this article
Devkota SP, Paudel A. New Delhi Metallo-β-lactamase among clinical isolates in Nepal. Edorium J Microbiol 2018;4:100010M08SD2018.ABSTRACT
Background: Incidence and drug resistance of the isolates producing New Delhi Metallo- β-lactamase (NDM) has been increasing throughout the world. Nepal is located between India and China from where abundant reports of NDM detection has been reported in recent days but in Nepal, very few studies are there. Hence this review was done to know the status of NDM in Nepal and its various characteristics.
Method: Research articles reporting NDM producing isolates from Nepal published before June 2018 were searched using various scientific databases using specific keywords and required data were extracted.
Result: According to the reviewed articles, NDM was prevalent in E.coli, Klebsiella, Acinetobacter, Pseudomonas, Providencia and other members of Enterobacteriaceae in Nepal. Seven variants of this gene were reported including NDM-1, NDM-3, NDM-4, NDM-7, NDM- 8, NDM-12, and NDM-13. These pathogens were not susceptible to almost all routine drugs due to co-existence of NDM gene with the wide variety of other drug resistance genes like Carbapenemases, ESBL, 16S rRNA methylases etc. Most of them were sensitive only to colistin, tigecycline, and fosfomycin. Some of these isolates were extremely drug-resistant as well as resistant to colistin, one of the last resort drugs.
Conclusion: High drug resistance of these isolates and their distribution among major human bacterial pathogens necessitates the prompt surveillance of these pathogens throughout the country, monitoring their resistant patterns, judicious antibiotic use, screening of these isolates from clinical, food, and environmental isolates and development of appropriate control measures.
Keywords: Clinical isolates, Nepal, New Delhi Metallo-β-lactamase
INTRODUCTION
NDM is a class B Metallo-β-lactamase, which is disseminated worldwide in different bacterial isolates after its first appearance in 2009 [1]. These isolates are predominant in the Indian subcontinent as they have been reported from India, Pakistan, and Bangladesh [2]. Isolates carrying NDM are regarded as superbugs due to their high drug resistance, except few last line antibiotics like colistin and tigecycline and dissemination of such isolates pose significant threat to global public health [3]. NDM has been detected among a variety of antibioticresistant gram-negative pathogens isolated from health care and community setting of Pakistan and India, as well from more than 15 countries of the globe [4]. NDM producing bacterial isolates are equipped with minimum one copy of partial or full ISAba125 gene which acts as a powerful promoter of this gene and NDM producing members of Enterobacteriaceae are related with travel to South Asia in many cases [5]. This Metallo-β-lactamase is a great concern as the gene for this enzyme is placed in the highly mobile genetic segment. In addition to this, it has a highly complex and possibly uncertain mode of dissemination than Klebsiella pneumoniae carbapenemase [6]. There is a simultaneous spread of NDM and emergence of its variants as indicated by reports of 10 NDM variants already. Some NDM variants like NDM-4 and NDM-5 pose higher carbapenemase activity compared to NDM-1 wild type, leading to increased resistance to β-lactam drugs and hence there is a demand of surveillance of NDM as well as its variants to know the epidemic and β-lactam resistance by these isolates [7]. Zinc ions are necessary for NDM to breakdown β-lactam drugs, as a result, its activity can be impeded by metal chelators i.e EDTA while its activity is not hindered by all known inhibitors of β-lactamase i.e tazobactam, sulbactam and clavulanate [8]. In this decade, NDM gene has also been detected from Nepal [9],[10],[11],[12],[13],[14],[15],[16],[17],[18],[19],[20].
Study of drug-resistance genes and their mechanism among clinical isolates are very rare in Nepal. Hence, the actual prevalence of ESBL, carbapenemase and other resistance genes among pathogens are not well known. Among the resistance genes, NDM is one of the significant genes as its high level of resistance and mode of rapid spread. The data regarding the status of NDM in Nepal is not available till now. Hence, this review was carried out to access the current scenario of NDM in Nepal and its various aspects. To our knowledge, it is the first review article regarding this highly notorious gene in Nepal.
MATERIALS AND METHODS
Literature search
A systematic literature search was carried out for New Delhi Metallo-β-lactamase in Nepal from various electronic databases (Medline via PubMed and Embase) published till May 2018. Original research articles available in English indicating NDM detection using molecular methods from Nepal were only included in this study (Figure 1). Following keywords were used during literature search in title and abstract of the articles: NDM, NDM variants, Nepal, gram-negative isolates, Multi-drug resistant, incidence.
Inclusion and exclusion criteria
Research articles fulfilling following criteria in abstract and/ or text were included in the study; i) detected NDM gene from gram-negative pathogens isolated in Nepal, ii) used valid molecular methods for the NDM gene detection, iii) provided various characteristics of NDM producers like isolate producing NDM, duration of study, NDM variants, study site and antibiotic susceptibility profile. Similarly, the exclusion criteria were; i) lacked the valid molecular method of NDM detection, ii) lacked various characteristics of NDM producers, iii) review articles, iv) duplicate articles of the same study, v) articles not in English language, vi) meta-analysis, and vii) abstract only articles.
Data extraction
Following variables were extracted from the selected studies: isolate producing NDM, source specimen of the isolates, duration of a study, the presence of other resistant gene, NDM variants, antibiotic susceptibility profile and study site. Two reviewers independently reviewed the selected articles fairly and discussions were made to settle discrepancies between two reviewers.
RESULT
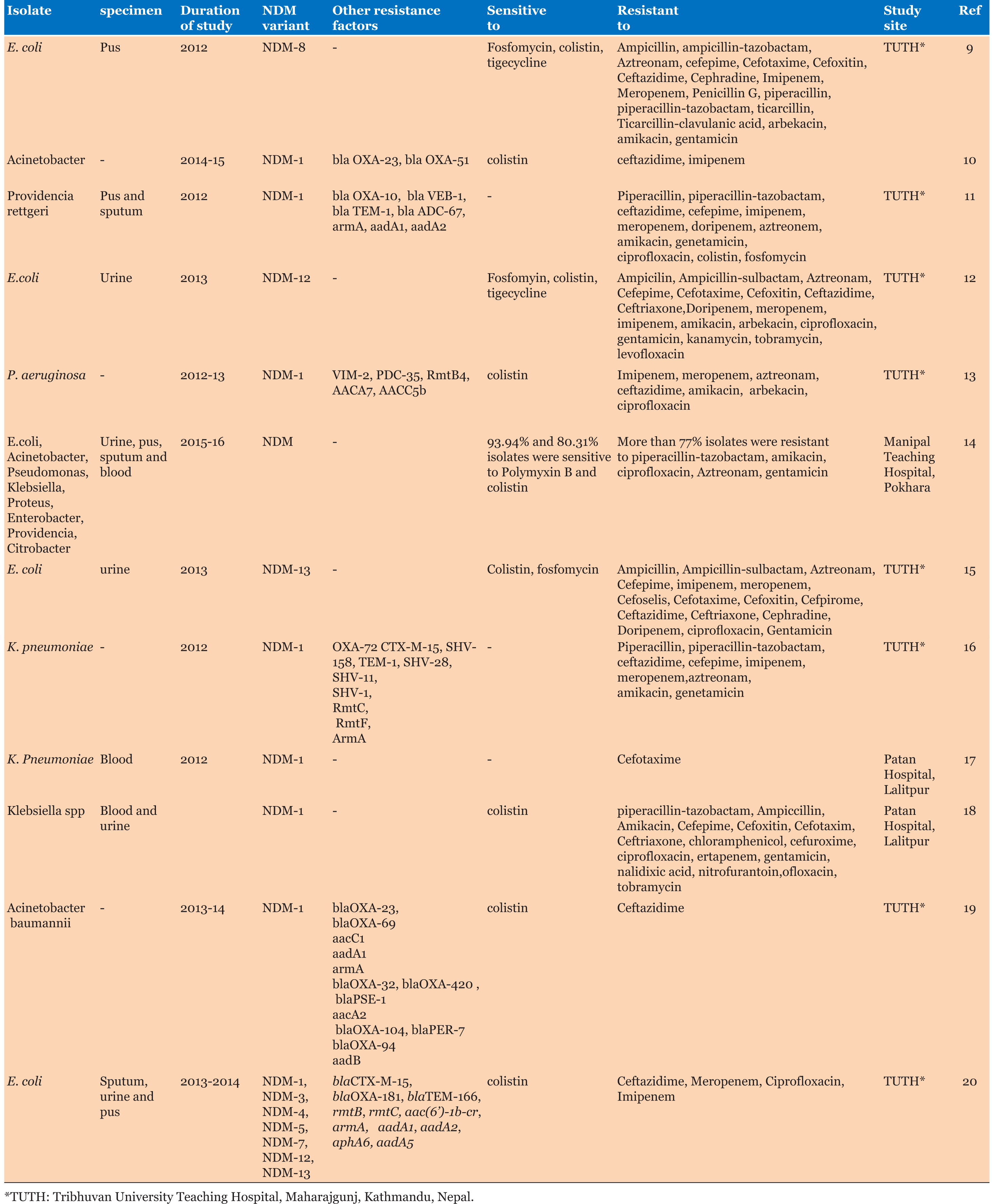
NDM producers were detected in members of Enterobacteriaceae (mainly E.coli and Klebsiella) and non-fermenting gram-negative isolates. These isolates have been reported from various clinical specimens after 2012 in Nepal. Most of the isolates were highly drug-resistant as they were resistant to almost all members of antibiotics related to different classes. Some of these isolates also produced various other resistant determinants leading to increased resistance to nearly all tested antibiotics as indicated by increased MIC values [11],[12]. They were also not affected by a combination of beta-lactam and beta-lactamase inhibitors like ampicillintazobactam piperacillin-tazobactam and ampicillinsulbactam (Table 1).
If we compare the distribution and other characteristics of NDM positive clinical isolates between Nepal and India, very similar results can be seen. In India, NDM has been reported in Acinetobacter baumannii [21], E.coli, Klebsiella spp, Enterobacter spp [22],[23], E.coli, Klebsiella spp, Citrobacter spp, Proteus spp, Serratia spp and Acinetobacter spp [24], Pseudomonas aeruginosa [25],[26], Klebsiella pneumoniae [27], E.coli [28] and E.coli, and Klebsiella pneumoniae [29]. Most common variants of NDM were NDM-1 [21],[22],[23],[24],[25] while other variants like NDM-4, NDM-5, and NDM-7 were also detected [28]. Indian NDM bearing isolates were also very resistant to routine antibiotics including imipenem, meropenem, gentamicin, amikacin, netlimicin, and tobramycin [21], ertapenem, aztreonam, and cefepime [25], ceftazidime, cefotaxime, ciprofloxacin, and tigecycline [27], ampicillin, ampicillin-sulbactam, cephalothin, cefoperazone, cefazolin, and clarithromycin [28] and piperacillintazobactam, cefpirome, and minocycline [29]. Such high drug resistance was attributed due to co-production of NDM genes with blaOXA [21],[22],[23], blaKPC-2 [25], blaVIM and blaIMP [26], blaKPC-2, blaSHV-12, blaCTX-M-15, blaTEM-1, rmtB and blaOXA-1 [27] and blaCTX-M-15, tem, aac6’, qnrB, qnrS, tetA, tetC, sul1, sul2, dfr and strA [28] genes. These isolates were sensitive, only to colistin or tigecycline or their combination [21],[27],[28],[29].
All these facts conclude that NDM producing isolates of these two nations are comparable in many aspects.
DISCUSSION
Based on the review, NDM gene was reported in Nepal for the first time in 2012 among E.coli, Providencia rettgeri, Pseudomonas and Klebsiella pneumoniae. This gene has been detected in other various gram-negative isolates from the variety of clinical samples (Table 1).
There is a continuous increase in prevalence and spread of this gene in the world [6]. This increase in the occurrence of these isolates may be due to use of antibiotics recklessly, lack of proper training to laboratory staffs, inadequate administration of prophylactic sanitary procedures and absence of policies to contain hospital infections [30]. Many factors play role in its worldwide spread including its dissemination among various bacterial pathogens as well as in plasmids of the different type. However, other factors like epidemiological and environmental factors assisting its globalization and responsible genetic mechanism are not known [4]. The plasmid carrying NDM gene is autotransmissible and versatile which may be the probable reason for its widespread dissemination among various microorganisms [31].
Many NDM producers were also positive for many other resistance genes including CTM-M, OXA, SHV, TEM, VIM, VEB, armA etc conferring resistance to the majority of members of different classes of antibiotics including penicillins, cephems, monobactams, carbapenems, aminoglycosides, and quinolones. Coexistence of various other resistant determinants is common among these isolates including ESBL genes, carbapenemases etc [4],[32],[33],[34],[35]. Co-production of such resistance gene is a matter of concern as it left the majority of available antibiotics ineffective. Isolates producing the variety of drug resistance genes in addition to NDM were extremely drug-resistant as found in this study. Where the majority of the isolates were equipped with various carbapenemase genes causing carbapenem resistance, beta-lactamase genes causing almost all beta-lactams ineffective, 16S rRNA methylase and aminoglycoside acetyl/adenylyltransferase causing high resistance to aminoglycosides.
Very few treatment options were available for the isolates producing this gene in Nepal as most of them were sensitive only to colistin, fosfomycin, and tigecycline. However, there is a rise of resistance against colistin among these isolate from Nepal [11],[14] and other countries [36] [37] indicating the possibility of pan-drug resistance in these isolates. Similarly, the study of Li et al has reported tigecycline and colistin were the only treatment option for NDM positive E. coli and K. pneumoniae isolates [3]. Likewise, there are various reports of susceptibility of these isolates only to last line drugs [8],[35]. Hence, there is an urgent need to search for combination drugs that can treat such isolates effectively as new drug discovery takes a long time. There are number of studies reporting the effectiveness of combined therapy against such isolates: rifampin-meropenem-colistin combination was effective against VIM and NDM producing K. pneumoniae isolates [38], amikacin and colistin against NDM-5 and MCR-1 positive E. coli [39].
Screening of these isolates in communities and healthcare settings is very crucial for their treatment and proper control as there is a report of the hospital outbreak of NDM positive K. pneumoniae causing high mortality (75%) among septicemic children admitted at Patan Hospital, Lalitpur, Nepal in 2012 [17]. Early detection, proper training of the laboratory staffs and regular screening of hospital environment and equipments for such pathogens can prevent such high mortality in addition to prevention of their spread to various hospital wards. Various factors associated with these isolates which make them precarious are i) most plasmids bearing NDM are capable of rearrangement and are mobile posing possibility of transmission to the variety of bacterial pathogens, ii) lack of standard phenotypic methods for their regular detection, iii) probability of high prevalence among unknown symptomless carriers, iv) absence of an effective treatment option among available antibiotics [40].
Similarities between various characteristics of NDM producers between India and Nepal may be due to the similar type of health care practice, self medication, and reckless use of antibiotics. In addition to this, the source of these isolates may have same origin as there is huge flow of people between these two nations as medical tourists, workers, and pilgrims. Such visits may have contributed to unnoticed transmission of such pathogens between these two adjacent countries as there are evidences of transmission of NDM positive isolates as a result of medical tourism [41] and mass movement of pilgrims [42] and most reported cases are related to visit of South Asian countries.
Study of these isolates is focused primarily on Kathmandu valley which is not sufficient to know the status of these pathogens in this country. Hence, there is imminent need of surveillance study of these isolates throughout the country among clinical, asymptomatic carriers, and environmental samples as various studies have reported their detection from carriers [43],[44] and environmental isolates [31],[45]. As drug-resistant isolates in intestine and environment may act as a potential source of human infection, hence multisector studies and implementing suitable control measures are the only possible ways of their management. International collaboration is very crucial for the control of this global public health challenge posed by drug resistance as this is a great problem to overcome [2]. NDM producing gramnegative isolates are present among clinical isolates in Nepal. This study is significant as most of these isolates were highly drug resistant due to the co-production of various resistant determinants.
CONCLUSION
Regular screening, proper training to health practitioners, development of simple as well as effective phenotypic methods for their detection and prudent antibiotic use are crucial to contain their spread. Further study about these pathogens is necessary in the Indian subcontinent as it is considered a major reservoir area for these superbugs.
REFERENCES
1.
Qu H, Wang X, Ni Y, et al. NDM-1-producing Enterobacteriaceae in a teaching hospital in Shanghai, China: IncX3-type plasmids may contribute to the dissemination of blaNDM-1. Int J Infect Dis 2015 May;34:8–13. [CrossRef]
[Pubmed]

2.
Johnson AP, Woodford N. Global spread of antibiotic resistance: The example of New Delhi metallo-ß-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 2013 Apr;62(Pt 4):499–513. [CrossRef]
[Pubmed]

3.
Li T, Wang Q, Chen F, et al. Biochemical characteristics of New Delhi metallo-ß-lactamase-1 show unexpected difference to other MBLs. PLoS One 2013 Apr 12;8(4):e61914. [CrossRef]
[Pubmed]

4.
Rasheed JK, Kitchel B, Zhu W, et al. New Delhi metallo-ß-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 2013 Jun;19(6):870–8. [CrossRef]
[Pubmed]

5.
Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother 2014 Jul;58(7):4211–3. [CrossRef]
[Pubmed]

6.
Bonomo RA. New Delhi metallo-ß-lactamase and multidrug resistance: A global SOS? Clin Infect Dis 2011 Feb 15;52(4):485–7. [CrossRef]
[Pubmed]

7.
Huang L, Hu X, Zhou M, et al. Rapid detection of New Delhi metallo-ß-lactamase gene and variants coding for carbapenemases with different activities by use of a PCR-based in vitro protein expression method. J Clin Microbiol 2014 Jun;52(6):1947–53. [CrossRef]
[Pubmed]

8.
Ranjan A, Shaik S, Mondal A, et al. Molecular epidemiology and genome dynamics of New Delhi metallo-ß-lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob Agents Chemother 2016 Oct 21;60(11):6795–805. [CrossRef]
[Pubmed]

9.
Tada T, Miyoshi-Akiyama T, Dahal RK, et al. NDM-8 metallo-ß-lactamase in a multidrug-resistant Escherichia coli strain isolated in Nepal. Antimicrob Agents Chemother 2013 May;57(5):2394–6. [CrossRef]
[Pubmed]

10.
Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of blaOXA-23 and blaNDM-1 genes of Acinetobacter baumannii isolated from Nepal: Antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control 2017 Feb 7;6:21. [CrossRef]
[Pubmed]

11.
Tada T, Miyoshi-Akiyama T, Dahal RK, et al. NDM-1 metallo-ß-lactamase and ArmA 16S rRNA methylase producing Providencia rettgeri clinical isolates in Nepal. BMC Infect Dis 2014 Feb 3;14:56. [CrossRef]
[Pubmed]

12.
Tada T, Shrestha B, Miyoshi-Akiyama T, et al. NDM-12, a novel New Delhi metallo-ß-lactamase variant from a carbapenem-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother 2014 Oct;58(10):6302–5. [CrossRef]
[Pubmed]

13.
Tada T, Shimada K, Satou K, et al. Pseudomonas aeruginosa clinical isolates in Nepal coproducing metallo-ß-lactamases and 16S rRNA methyltransferases. Antimicrob Agents Chemother 2017 Aug 24;61(9). [CrossRef]
[Pubmed]

14.
Devkota SP, Sharma S, Bhatta DR, Paudel A, Sah AK, Kandel BP. Prevalence of the blaNDM gene among metallo-ß-lactamase-producing Gram-negative isolates from western Nepal. J Glob Antimicrob Resist 2018 Mar;12:3–4. [CrossRef]
[Pubmed]

15.
Shrestha B, Tada T, Miyoshi-Akiyama T, et al. Identification of a novel NDM variant, NDM-13, from a multidrug-resistant Escherichia coli clinical isolate in Nepal. Antimicrob Agents Chemother 2015 Sep;59(9):5847–50. [CrossRef]
[Pubmed]

16.
Tada T, Miyoshi-Akiyama T, Dahal RK, et al. Dissemination of multidrug-resistant Klebsiella pneumoniae clinical isolates with various combinations of carbapenemases (NDM-1 and OXA-72) and 16S rRNA methylases (ArmA, RmtC and RmtF) in Nepal. Int J Antimicrob Agents 2013 Oct;42(4):372–4. [CrossRef]
[Pubmed]

17.
Chung The H, Karkey A, Pham Thanh D, et al. A high-resolution genomic analysis of multidrug-resistant hospital outbreaks of Klebsiella pneumoniae. EMBO Mol Med 2015 Mar;7(3):227–39. [CrossRef]
[Pubmed]

18.
Amatya P, Joshi S, Shrestha S. Outbreak of extended spectrum beta lactamase producing Klebsiella species causing neonatal sepsis at patan hospital in Nepal. Journal of Patan Academy of Health Sciences 2014;1(1):20–2. [CrossRef]
[Pubmed]

19.
20.
Shrestha B, Tada T, Shimada K, et al. Emergence of various NDM-type-metallo-ß-lactamase-producing Escherichia coli clinical isolates in Nepal. Antimicrob Agents Chemother 2017;61(12):e01425–17. [CrossRef]
[Pubmed]

21.
Karthikeyan K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumannii from India. J Antimicrob Chemother 2010 Oct;65(10):2253–4. [CrossRef]
[Pubmed]

22.
Manohar P, Shanthini T, Ayyanar R, et al. The distribution of carbapenem- and colistin-resistance in Gram-negative bacteria from the Tamil Nadu region in India. J Med Microbiol 2017 Jul;66(7):874–83. [CrossRef]
[Pubmed]

23.
24.
Naim H, Rizvi M, Azam M, et al. Alarming emergence, molecular characterization, and outcome of blaNDM-1 in patients infected with multidrug-resistant Gram-negative bacilli in a tertiary care hospital. J Lab Physicians 2017 Jul–Sep;9(3):170–6. [CrossRef]
[Pubmed]

25.
Paul D, Dhar Chanda D, Maurya AP, et al. Co-Carriage of blaKPC-2 and blaNDM-1 in Clinical Isolates of Pseudomonas aeruginosa Associated with Hospital Infections from India. PLoS One. 2015 Dec 29;10(12):e0145823. [CrossRef]
[Pubmed]

26.
Mohanam L, Menon T. Coexistence of metallo-beta-lactamase-encoding genes in Pseudomonas aeruginosa. Indian J Med Res 2017 Jul;146(Supplement):S46–S52. [CrossRef]
[Pubmed]

27.
Kumarasamy K, Kalyanasundaram A. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother 2012 Jan;67(1):243–4. [CrossRef]
[Pubmed]

28.
Ranjan A, Shaik S, Mondal A, et al. Molecular epidemiology and genome dynamics of New Delhi metallo-ß-lactamase-producing extraintestinal pathogenic Escherichia coli strains from India. Antimicrob Agents Chemother 2016 Oct 21;60(11):6795–805. [CrossRef]
[Pubmed]

29.
Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis 2010 Sep;10(9):597–602. [CrossRef]
[Pubmed]

30.
Dadashi M, Fallah F, Hashemi A, et al. Prevalence of blaNDM1–producing Klebsiella Pneumoniae in Asia: A Systematic review and meta-analysis. Journal des Anti-infectieux 2017;19(2):58–65. [CrossRef]

31.
Islam MA, Islam M, Hasan R, et al. Environmental spread of New Delhi metallo-ß-lactamase-1-producing multidrug-resistant bacteria in Dhaka, Bangladesh. Appl Environ Microbiol 2017 Jul 17;83(15). [CrossRef]
[Pubmed]

32.
Zhou G, Guo S, Luo Y, et al. NDM-1-producing strains, family Enterobacteriaceae, in hospital, Beijing, China. Emerg Infect Dis 2014 Feb;20(2):340–2. [CrossRef]
[Pubmed]

33.
Xie L, Dou Y, Zhou K, et al. Coexistence of blaOXA-48 and truncated blaNDM-1 on different plasmids in a Klebsiella pneumoniae isolate in China. Front Microbiol 2017 Feb 2;8:133. [CrossRef]
[Pubmed]

34.
Zhang X, Lou D, Xu Y, et al. First identification of coexistence of blaNDM-1 and blaCMY-42 among Escherichia coli ST167 clinical isolates. BMC Microbiol 2013 Dec 5;13:282. [CrossRef]
[Pubmed]

35.
Zhao JY, Zhu YQ, Li YN, et al. Coexistence of SFO-1 and NDM-1 ß-lactamase genes and fosfomycin resistance gene fosA3 in an Escherichia coli clinical isolate. FEMS Microbiol Lett 2015 Jan;362(1):1–7. [CrossRef]
[Pubmed]

36.
Zhong LL, Zhang YF, Doi Y, et al. Coproduction of MCR-1 and NDM-1 by Colistin-Resistant Escherichia coli Isolated from a Healthy Individual. Antimicrob Agents Chemother 2016 Dec 27;61(1). [CrossRef]
[Pubmed]

37.
Lomonaco S, Crawford MA, Lascols C, et al. Resistome of carbapenem- and colistin-resistant Klebsiella pneumoniae clinical isolates. PLoS One 2018 Jun 8;13(6):e0198526. [CrossRef]
[Pubmed]

38.
Tängdén T, Hickman RA, Forsberg P, Lagerbäck P, Giske CG, Cars O. Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time-kill experiments. Antimicrob Agents Chemother 2014;58(3):1757–62. [CrossRef]
[Pubmed]

39.
Zhou YF, Tao MT, Feng Y, et al. Increased activity of colistin in combination with amikacin against Escherichia coli co-producing NDM-5 and MCR-1. J Antimicrob Chemother 2017 Jun 1;72(6):1723–30. [CrossRef]
[Pubmed]

40.
Khan AU, Maryam L, Zarrilli R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-ß-lactamase (NDM): A threat to public health. BMC Microbiol 2017 Apr 27;17(1):101. [CrossRef]
[Pubmed]

41.
Chen LH, Wilson ME. The globalization of healthcare: Implications of medical tourism for the infectious disease clinician. Clin Infect Dis 2013 Dec;57(12):1752–9. [CrossRef]
[Pubmed]

42.
Ahammad ZS, Sreekrishnan TR, Hands CL, Knapp CW, Graham DW. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the upper ganges river. Environ Sci Technol 2014;48(5):3014–2. [CrossRef]
[Pubmed]

43.
Perry JD, Naqvi SH, Mirza IA, et al. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother 2011 Oct;66(10):2288–94. [CrossRef]
[Pubmed]

44.
Berrazeg M, Diene S, Medjahed L, Parola P, Drissi M, Raoult D, Rolain J. New Delhi Metallo-beta-lactamase around the world: An eReview using Google Maps. Euro Surveill 2014 May 22;19(20). pii: 20809.
[Pubmed]

45.
Mantilla-Calderon D, Jumat MR, Wang T, Ganesan P, Al-Jassim N, Hong PY. Isolation and Characterization of NDM-Positive Escherichia coli from Municipal Wastewater in Jeddah, Saudi Arabia. Antimicrob Agents Chemother 2016 Aug 22;60(9):5223–31. [CrossRef]
[Pubmed]

SUPPORTING INFORMATION
Author Contributions
Surya Prasad Devkota - Substantial contributions to conception and design, Acquisition of data, Analysis of data, Interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published
Ashmita Paudel - Substantial contributions to conception and design, Acquisition of data, Analysis of data, Interpretation of data, Drafting the article, Revising it critically for important intellectual content, Final approval of the version to be published
Guaranter of SubmissionThe corresponding author is the guarantor of submission.
Source of SupportNone
Data AvailabilityAll relevant data are within the paper and its Supporting Information files.
Conflict of InterestAuthors declare no conflict of interest.
Copyright© 2018 Surya Prasad Devkota et al. This article is distributed under the terms of Creative Commons Attribution License which permits unrestricted use, distribution and reproduction in any medium provided the original author(s) and original publisher are properly credited. Please see the copyright policy on the journal website for more information.